Salmonella Rapid Culture Method
Salmonella is a Gram-negative, rod-shaped, motile bacterium with a widespread occurrence in animals, especially in poultry and swine. Environmental sources of the organism include raw meat, raw poultry and raw seafood, to name only a few. Detection is critical as is the most frequently reported cause of foodborne illness (40,000–50,000 cases reported annually), and the infectious dose can be as low as 1–10 cells.
Current Methods
Current testing methods (FDA/BAM) involve a pre-enrichment step followed by a second selective enrichment step, followed by plating (which can include secondary plating). This process takes 72–96 hours (3–4 days), depending on the food sample being analyzed, and must be followed by biochemical/serological confirmation of Salmonella contamination, increasing the time to results to up to 5 days.
As indicated above, two enrichment steps are typically needed to recover Salmonella since the bacteria are usually present in very low numbers and may be stressed or damaged from food processing methods. These enrichment steps are general enrichments and are designed to cultivate any microorganisms that may be present in the food sample, potentially masking the presence of very low numbers of true pathogens.
In addition, the secondary enrichment media, modified Rappaport Vassiliadis broth, must be prepared from its individual raw materials to ensure sufficient recovery of any contaminating microorganisms.
Rapid Confirmation
To address the need for a rapid, accurate test for confirmation of Salmonella contamination in food products, a new method, the Salmonella Rapid Culture Method, has been developed. This method involves a single enrichment in ONE Broth-Salmonella, followed by plating on Brilliance™ Salmonella chromogenic medium, taking - less than 2 days to generate an initial result. Thermo Scientific recently received AOAC-RI certification for this method (License no. 120802).
ONE Broth-Salmonella is a highly nutritious enrichment broth containing a specific growth promoter to ensure excellent recovery of stressed and damaged Salmonella cells, while inhibiting the growth of competing microorganisms. This effective medium allows enrichment to be performed in a single 18–24 hour incubation, eliminating the need for a secondary enrichment.
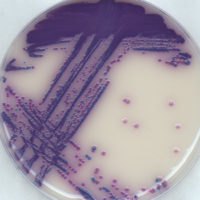
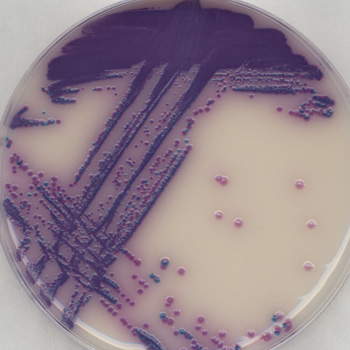
Following enrichment, the sample is plated onto chromogenic culture media. Chromogens within the medium enable differentiation of Salmonella colonies (bright purple) from any remaining organisms that are able to grow, such as Klebsiella (blue colonies) and Enterobacter (clear colonies to no growth), thus reducing the number of false positives that require confirmation.
Testing Results
Fresh Salmonella cultures were prepared from frozen stocks using standard enrichment media [tryptic soy broth (TSB), brain heart infusion (BHI)] or ONE Broth-Salmonella) and incubated for 18–24 hours at the appropriate temperatures, according to manufacturers’ or guideline instructions.
Raw ground beef, ground chicken, lettuce, shrimp and egg shells were obtained from local suppliers. Food samples were inoculated with low [~1 colony forming unit (CFU)/25g] or high (~1.1 CFU/g) microorganism amounts. The final inoculated concentrations were confirmed by MPN (most probable number). Side-by-side samples (both inoculated with micro-organisms and uninoculated negative controls) were compared using the Rapid Salmonella method and by the appropriate USDA/FSIS and FDA-BAM reference methods for each matrix.
A total of 102 Salmonella serovars/ strains were obtained and tested as indicated above. Almost all (98 of 102) of these strains produced a positive purple-colored colony. In addition, a total of 30 species not belonging to the genus Salmonella were analyzed in the same manner. All these strains showed atypical (non-purple colonies) or no growth.
Summary
This study demonstrated that the rapid method described above detected almost all serovars of Salmonella when spiked at levels from low to high in food sources in which Salmonella contamination has been observed. Since 98/102 serovars produced the “typical” purple colonies, it is highly unlikely that a specific serovar would go undetected and lead to an outbreak of salmonellosis (the serovars not detected are not typically found in food). By screening such large numbers of serovars/species, Thermo Scientific is helping food companies to feel secure knowing that food is free from Salmonella contamination when tested using this methodology.
In addition, this study demonstrated that it is possible to discriminate between Salmonella species and other selected non-Salmonella organisms, demonstrating the method’s selectivity. It also ensures that samples contaminated with non-Salmonella organisms do not demonstrate a false-positive color development, allowing the proper identification of food pathogens present in the food being tested. This will ensure a safe food supply free of Salmonella while preventing the waste from discarding any false-positive food lots.
This robust method makes rapid identification of pathogens in food products faster and easier, reducing time to market and eliminating the risk of food recalls.
www.remel.com/salmonella
Looking for a reprint of this article?
From high-res PDFs to custom plaques, order your copy today!